Summary
Zip Nucleic Acids (ZNA®) are a novel class of modified oligonucleotides enhancing hybridization properties of nucleic acids. ZNA® are oligonucleotides conjugated to cationic units. ZNA® show increased affinity for their targets by reducing the electrostatic repulsion between nucleic acid strands. ZNA® increase both the binding efficiency and the kinetic, and stabilize the hybridization to their target. These properties make ZNA® promising powerful tools for molecular biology and diagnostic applications.
When used as PCR primers ZNA® increase the amplification efficiency and the specificity and are particularly efficient in A-T rich sequences or for low abundant targets.
When used as probes in PCR assays, ZNA® increase the sensitivity and specificity of detection making them optimal tools for SNP discrimination or miRNA detection. They are easy to predict and design without affecting the hybridization specificity between complementary sequences. In addition, they are easy to manufacture due to their diblock conjugate structure and the novel phosphoramidite chemistry that has been developed by Polyplus-transfection® is fully adapted to the automated oligonucleotide synthesis.
Description
What are ZNA®?
ZNA® are formed by conjugating spermine derivatives, as cationic moieties (Z units), to oligonucleotides. Consequently, ZNA® increase the affinity of oligonucleotides for their target by decreasing the electrostatic repulsions due to the polyanionic nature of nucleic acids. ZNA® are made on a standard oligonucleotide synthesizer using the phosphoramidite chemistry, providing fast, and cost effective production (Pons et al., 2006; Voirin et al., 2007).
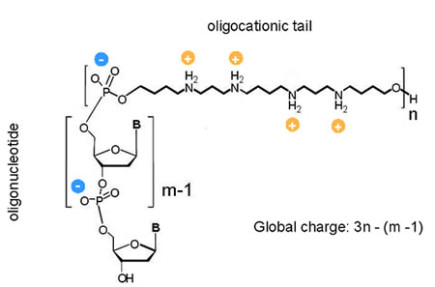
Structure of a ZNA molecule
NA® are versatile as the number of Z units can be chosen and placed either internally or at the end (5’, 3’) of the oligonucleotide depending on the application. By selecting the number of cationic units, the global charge of the ZNA® molecule can be modulated. In addition, many modifications such as dyes and quenchers can be added.
What are the properties of ZNA®?
Easy to design by modulating the Tm of oligos
The melting temperature (Tm) of ZNA® increases significantly and linearly with the number of cationic units grafted on the oligonucleotide, providing a convenient means for fine tuning hybridization temperatures (Noir et al., 2008).
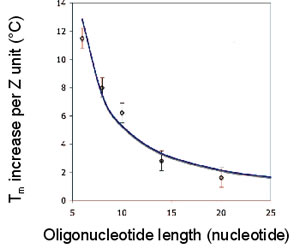
Tm vs ZNA Oligonucleotide length
The Tm increases linearly with the number of grafted Z units, irrespectively of the base sequence of the oligonucleotide and of the conjugation site of the cationic Z units (3’ or 5’). The Tm increase per Z unit only depends on the length of the oligonucleotide.
The Tm of the ZNA® is then easily predictable, using a simple mathematical relation depending on the intrinsic DNA oligonucleotide Tm, on the length N of the oligonucleotide and on the number Z of cationic units Tm (ZNA) = Tm (DNA) + 36z/(N-3.2), (Noir et al., 2008).
Higher affinity, improved binding and faster kinetics
ZNA® was shown to facilitate the hybridization with a target sequence by clipping the complementary strands together like a zipper, hence their name Zip nucleic acids. ZNA® enhance the affinity for their target by accelerating the complementary sequence recognition. ZNA® increase the binding and stabilize the formed duplex by reducing electrostatic repulsion, thereby increasing the Tm.
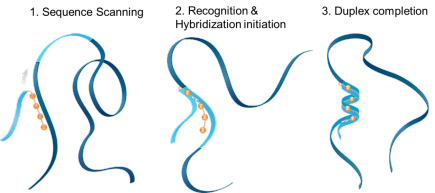
ZNA hybridization
Video: Watch ZNA® in action!
Applications
ZNA® as primers for PCR and RT-PCR
ZNA® primers can be used for PCR, quantitative PCR and RT-PCR (Reverse Transcription PCR), fast PCR, multiplex assays, A-T rich region amplification, or high-throughput PCR. ZNA® primers were found to drive PCR or RT-PCR reactions very efficiently with the following properties:
- Easy to design,
- High specificity,
- High sensitivity,
- Faster binding,
- High affinity (low concentration of primers),
- Efficient at high annealing temperature,
- Efficient in AT-rich regions,
- Circumvent [Mg2+] adjustment / variability,
- Improved RNA to cDNA conversion in RT-PCR,
- More accurate amplification of low abundant transcripts.
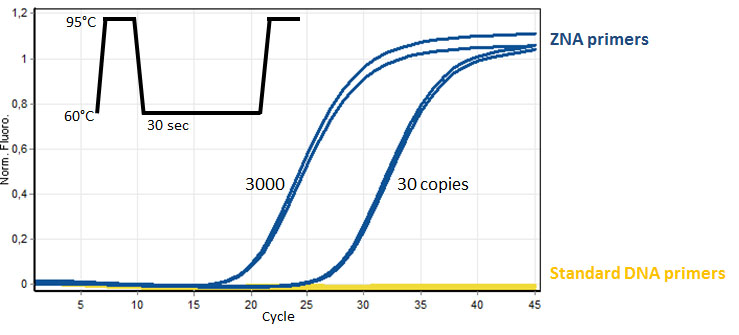
Efficient DNA amplification with AT-rich ZNA® primers (100 nM each, 70 and 80% AT, respectively) of target genomic HPV-16 (30 and 3000 copies) spiked in negative control DNA (10 ng). Comparatively standard DNA primers are not able to amplify the target genomic DNA in the same conditions. Please refer to Moreau et al., 2009.
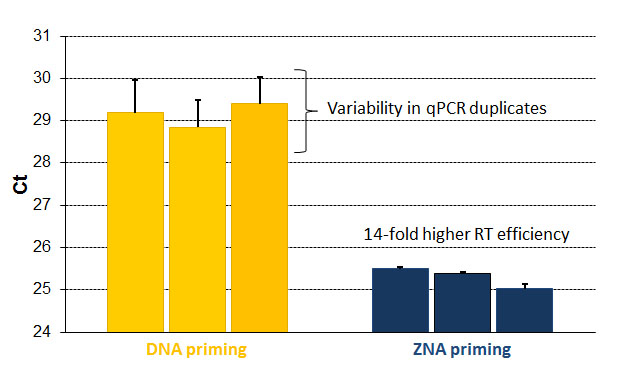
ZNA® primers improve the conversion of RNA to DNA of low abundant transcripts. Conversion comparison of the low expressed HMGA-2 transcripts to DNA obtained by RT of 200 ng of total RNA extracted form HeLa cells using 100 nM of standard DNA primer and ZNA® primer. Please refer to Moreau et al., 2009.
Other ZNA® primer applications
Other ZNA® primer applications include:
- Fast PCR,
- Multiplex assays,
- High-throughput PCR,
- Isothermal amplification.
ZNA® as dual-labeled probes for PCR
ZNA® dual-labeled probes improve performances of 5’ nuclease assays with the following properties:
- Flexible probe design,
- Higher Tm for short probes improving discrimination (SNP genotyping, miRNA detection),
- Improved quenching for long probes (low fluorescence background and increased signal-to-noise ratio),
- Earlier Cq values,
- Higher signal level.
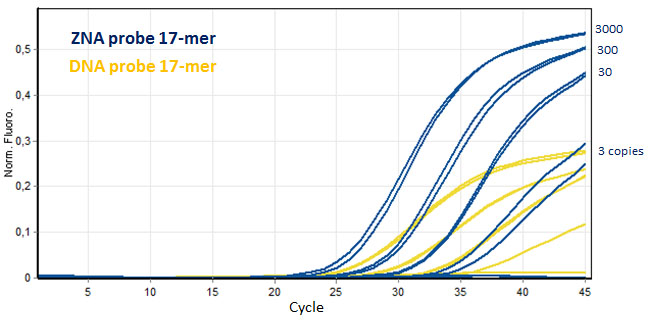
ZNA® improve the PCR detection of short dual labeled probes. Serial dilution amplifications of target genomic Factor V DNA (3000 to 3 copies) were more easily and efficiently detected using ZNA® short probe (17-mer, FAM-, BHQ-1 probe) and compared to its unmodified DNA probe counterpart. Please refer to Paris et al., 2010.
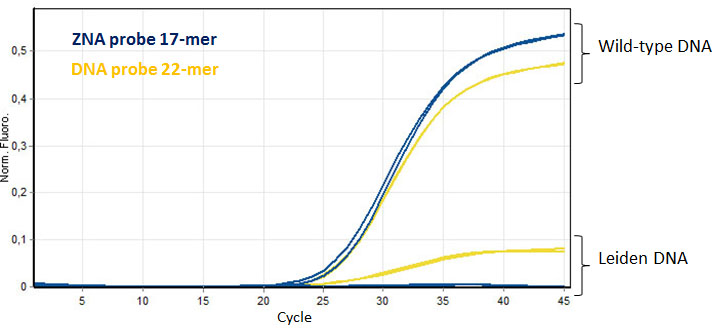
ZNA® short probe provide better SNP discrimination. Amplification of 10 ng of wild-type and Leiden genomic DNA were detected using a short ZNA® probe (17-mer) and compared to a longer unmodified DNA probe (22-mer). Please refer to Paris et al., 2010.
Other ZNA® probe applications
Other ZNA® probe applications include:
- miRNA detection (Kandemir et al., 2014 ; Tombiak et al., 2015),
- Detection of AT-rich sequences,
- PCR hybridization (non hydrolysable probes),
- In situ hybridization probes (Trevisan et al., 2012 ; Begheldo et al., 2013).
Citations
Moreau, V. et al. (2009) Zip nucleic acids (ZNAs): new high affinity oligonucleotides as potent primers for PCR and reverse transcription. Nucl. Acids Res., 37: e130.
Paris, C. et al. (2010) Zip nucleic acids are potent hydrolysis probes for quantitative PCR. Nucl. Acids Res., 38: e95.
Kandemir, H. et al. (2014) Evaluation of several micro RNA (miRNA) levels in children and adolescents with attention deficit hyperactivity disorder. Neuroscience Letters 2014, 580, 158-162.
Tombak, A. et al. (2015) MicroRNA expression analysis in patients with primary myelofibrosis, polycythemia vera and essential thrombocythemia. Indian J. Hematol. & Blood Transfusion 2015, DOI 10.1007/s12288-014-0492-z.
Begheldo, M. et al. (2013) Whole-mount in situ detection of microRNAs on Arabidopsis tissues using Zip Nucleic Acid probes. Anal Biochem, 434, 60-66.
Van Heuverswyn et al. (2016) Influence of primer & probe chemistry and amplification target on reverse transcription digital PCR quantification of viral RNA. Biomol Detect Quantif, 9, 20-28
Alayan et al. (2013) Differential expression of colon cancer associated transcript1 (CCAT1) along the colonic adenoma-carcinoma sequence. BMC Cancer, DOI: 10.1186/1471-2407-13-196
Find more publications on the Polyplus-transfection database by selecting ZNA from the « Molecules » picklist.
Intellectual Property, Partnering and licensing
ZNA® oligonucleotides, Spermine phosphoramidite and their use are the subject matter of (i) U.S. Patent No. 9090648, U.S. Divisional Patent Application No. 14/745,871, European Patent No. 1 973 927 and foreign equivalents entitled “Cationic oligonucleotides, automated methods for preparing same and their uses” and (ii) U.S. Patent No. 8,465,920, European Patent No. 2 225 393 and foreign equivalents entitled “Methods for hybridizing nucleic acids”.
The purchase of ZNA® oligonucleotides or Spermine Phosphoramidite does not include or carry an implied right or license for the buyer to use such products for commercial purposes. A license must be obtained directly from Polyplus-transfection for commercial use of these products, including but not limited to use in diagnostic procedures. For a commercial license to the ZNA® Intellectual Property, please visit our webpage dedicated to Licensing Opportunities and feel free to contact us .
Purchasing ZNA® or Spermine Phosphoramidite
Evaluation of the technology is easily available by purchase of the related research use products.
Custom ZNA® oligonucleotides are available from Metabion and TriLink BioTechnologies.
Spermine Phosphoramidite is available from Glen Research. More information in the Glen Reports « Spermine Phosphoramidite: A potent modification with many applications » and « Zip Nucleic Acids (ZNA®) are powerful cationic oligonucleotides for molecular biology, diagnostic and therapeutic applications ».
 |
………….. |  |
………….. |  |
