jetMESSENGER® is a designed for high mRNA transfection efficiency in primary cells, cancer cell lines, neurons, and stem cells.
We use cookies to help you navigate efficiently and perform certain functions. You will find detailed information about all cookies under each consent category below.
The cookies that are categorized as "Necessary" are stored on your browser as they are essential for enabling the basic functionalities of the site. ...
Necessary cookies are required to enable the basic features of this site, such as providing secure log-in or adjusting your consent preferences. These cookies do not store any personally identifiable data.
Functional cookies help perform certain functionalities like sharing the content of the website on social media platforms, collecting feedback, and other third-party features.
Analytical cookies are used to understand how visitors interact with the website. These cookies help provide information on metrics such as the number of visitors, bounce rate, traffic source, etc.
Advertisement cookies are used to provide visitors with customized advertisements based on the pages you visited previously and to analyze the effectiveness of the ad campaigns.
Authors: Alengo Nyamay’Antu, Fanny Prémartin, Thibaut Benchimol, Valérie Toussaint, Maxime Dumont, Patrick Erbacher
Introduction
The association of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated (Cas) nucleases is an innovative technology to generate gene knock-outs, introduce or delete sequences within the genome. As most of the microbial adaptive immune system relies on CRISPR-mediated defense against exogenous genetic elements, there is a diverse set of CRISPR/Cas systems available that consists of a guide RNA (gRNA) molecule that drives the endonuclease to a specific sequence within the genome to create a double-strand break.
Streptococcus pyogenes Cas9 (SpCas9) is the most commonly used Cas nuclease for site-specific gene targeting; there is a higher chance of finding its protospacer adjacent motif (PAM) 3’-NGG-5’ in the human genome. The successful use of Cas9 protein for specific genome editing in mammalian cells has opened Pandora’s box for engineered Cas9 mutants as well as other RNA-guided endonucleases. Among the other Cas RNA-guided endonuclease, acidaminococcus Cas12a protein also known as Cpf1 improves genome coverage for AT-rich regions.
Successful and efficient delivery of the gRNA and Cas protein pass the extracellular membrane into the cytosol is key to reaching high genome editing in mammalian cells. Focusing on the main CRISPR/Cas-mediated genome editing approach, there are different approaches to introduce gRNA and the Cas 9 protein in mammalian cells, including DNA- and RNA-based approaches (Fig. 1). DNA-free delivery systems in which Cas9 protein is delivered as mRNA is an alternative approach to improve genome editing efficiency in hard to transfect cells where DNA delivery into the nucleus is limited. In contrast to a plasmid DNA approach, delivery of Cas9 as mRNA leads to more transient and time-restricted Cas9 endonuclease activity, which can be beneficial when off-target nuclease activity is a concern.
Here, we present jetMESSENGER® our latest innovative reagent designed to co-deliver Cas9 mRNA and guide RNA. jetMESSENGER® transfection reagent achieves efficient delivery of CRISPR/Cas9 components in a wide variety of cells, as transcription to obtain a functional Cas9 protein within the cell is bypassed.
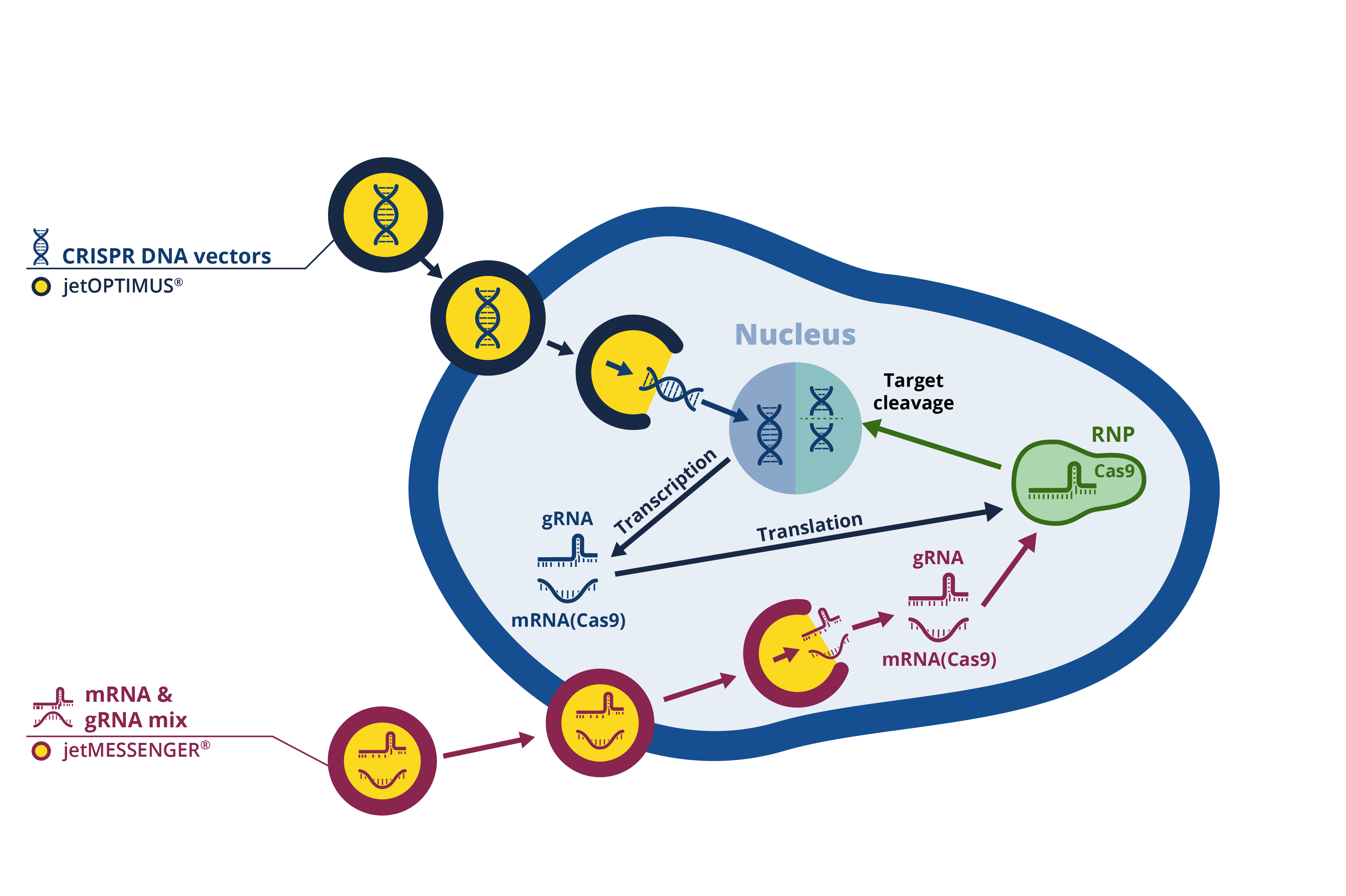
Fig. 1: in vitro transfection approaches for CRISPR experiments.
jetMESSENGER® transfection reagent is an innovative transfection reagent which efficiently delivers mRNA Cas9 and gRNA complexes to reach highest Cas9-mediated genome editing. During transfection optimization, we carefully optimized gRNA and Cas9 quantities to ensure highest percentage of indel (%) as a readout of genome editing efficiency. As shown in (Fig. 2),we demonstrate that jetMESSENGER® achieves above 50%of INDEL of the frequently used housekepping gene HPRT1 when using 50 ng of HPRT1 gRNA in combination with 100 ng of Cas9 mRNA. The increased % INDEL was signifcantly improved with up to 70% of INDEL when doubling HPRT1 gRNA quantity, with minimal impact on cell viability and cell morphology.
Fig. 2: Amazing genome editing efficiency obtained with jetMESSENGER® using RNA genome editing method. Cotransfections of 100 ng of Cas9 mRNA and 50 or 100 ng of HPRT1 gRNA were performed in HEK-293 cells with 0.3 µl of jetMESSENGER® reagent, per well of a 96-well plate. At 48 h post-transfection, genome editing was assessed by calculating the percentage (%) of INDEL using the T7 endonuclease method. The INDEL % was determined by using Genetools software (Syngene®).
There is a growing number of Cas-based strategies to address limitations of Cas9 in achieving specific and efficient genome editing. Cpf1, also known as Cas12 is used for its properties in targeting AT-rich regions for which Cas9 is less specific. As summarized in Table 2, Cpf1 structurally and functionally differs from Cas9 protein. On a structural level, Cpf1 and its corresponding gRNA are smaller in size then their equivalent in the CRISPR/Cas9 system: the gRNA is only composed of the crRNA sequence without the adjacent tracrRNA sequence which is required to drive the Cas9 protein to the targeted DNA sequence. On a functional level, the endonuclease activity of Cpf1 differs from that of Cas9 by cleaving genomic DNA in a staggered manner to create 5’ overhangs instead of 3’ blunt end cuts. The overhang cut generated by Cpf1 is expected to reduce occurrence of blunt end mutations, as well as increase the correct insertion rate of engineered sticky DNA sequences in knock-in studies.
Table 2: Structural and functional comparison of CRISPR/Cas9 and CRISPR/Cpf1 systems.
Conclusion
Successful delivery of the gRNA and the preferred Cas protein into cells is essential to guarantee the high genome editing efficiency that is required to generate new cell or animal models. The most efficient CRISPR-Cas DNA-free method is based on the direct delivery of gRNA and Cas protein as mRNA. mRNA transfection has noteworthy advantages compared to a plasmid-based approach: an increased control over Cas9 or Cpf1 endonuclease activity, reduced amount of off-target effects, and increased post-transfection cell viability. With our recently launched mRNA transfection reagent jetmESSENGER®, we now provide a full range of reagents forCas9 and Cpf1-based CRISPR approaches (Table 1).
Use the leading technology. Rely on Polyplus-transfection®.
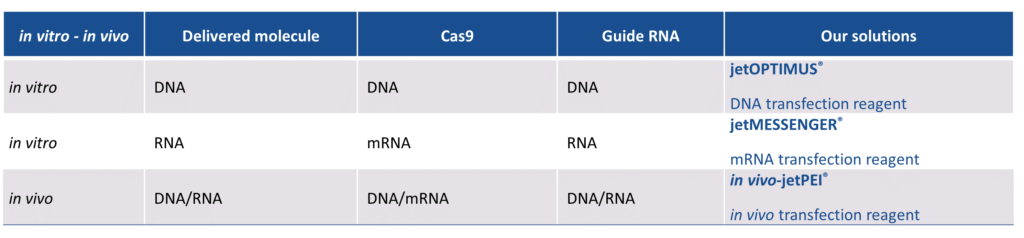
Table 3: Polyplus-transfection’s range of transfection reagents for CRISPR experiments.