jetPRIME® is a versatile DNA and siRNA transfection reagent that ensures high DNA transfection efficiency and excellent gene silencing.
Introduction
The use of the CRISPR/Cas9 technology in mammalian cells has recently emerged as a very convenient way to modify the cell genome at a specific locus. The CRISPR/Cas9 system is a prokaryotic immune mechanism that confers resistance to foreign genetic elements and consists of a DNA endonuclease Cas9 and a non-coding guide RNA (gRNA). The gRNA guides Cas9 to a specific complementary genomic locus to introduce a double-strand break.
The success of CRISPR genome editing is limited by the intracellular delivery and expression of Cas9 protein and gRNA. The most common strategy involves transient transfection into mammalian cells of one or several plasmids coding for Cas9, the specific gRNA and if needed the sequence to be inserted.
Here we show that efficient plasmid delivery with low DNA amount and low toxicity can be achieved with the Polyplus-transfection® reagent jetPRIME® for successful CRISPR/Cas9-mediated gene knock-out, as illustrated for two genes encoding essential proteins involved in DNA replication (PCNA and CDC45).
Material & methods
Cell culture and transfection
HeLa cells were maintained in Dulbecco’s modified Eagle’s tissue culture medium supplemented with 10% heat-inactivated fetal calf serum at 37°C in a humidified 5% CO2 atmosphere. All transfections were performed in 12-well plates (Greiner Bio One) at a plating density of 105 cells per well 24 h before transfection. Transfection of HeLa cells was performed with jetPRIME® (Polyplus-transfection®) using 0.8 µg DNA and 1.6 µL reagent per well.
CRISPR/Cas9 assay
Cells were transfected with a single plasmid encoding for Cas9, a gRNA sequence to target either PCNA or CDC45 gene, together with a puromycin resistance cassette (Fig 1). Two days after transfection, cells harboring the Cas9-encoding vector were selected using 2 µg/mL puromycin for 4 days. HeLa cells were observed with optical microscopy to verify the proper knockout of the PCNA or CDC45 gene (Fig 2A).
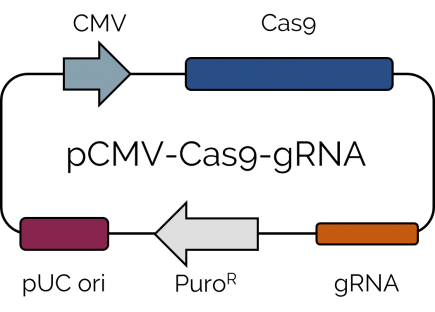
Fig 1. Schematic map of the pCMV-Cas9-gRNA plasmid.
Results
Blockage of DNA synthesis leads to extensive DNA damage such as double-strand breaks (DSBs). It has been reported that cells subjected to inhibition of DNA duplication are affected by phenotypical changes such as nuclear size and cytoplasm enlargement before subsequent cell death. Moreover, proteins involved in DNA synthesis like PCNA (processivity factor) and CDC45 (helicase cofactor) have been identified as markers of replication stress (Desplancq et al., 2016; Srinivasan et al., 2013).
To investigate whether knock-out of PCNA or CDC45 gene can be achieved using the CRISPR/Cas9 method, HeLa cells were transiently transfected with pCMV-Cas9-gRNA following the jetPRIME® protocol. Upon puromycin selection, transfected cells were observed with optical microscopy (Fig 2B).
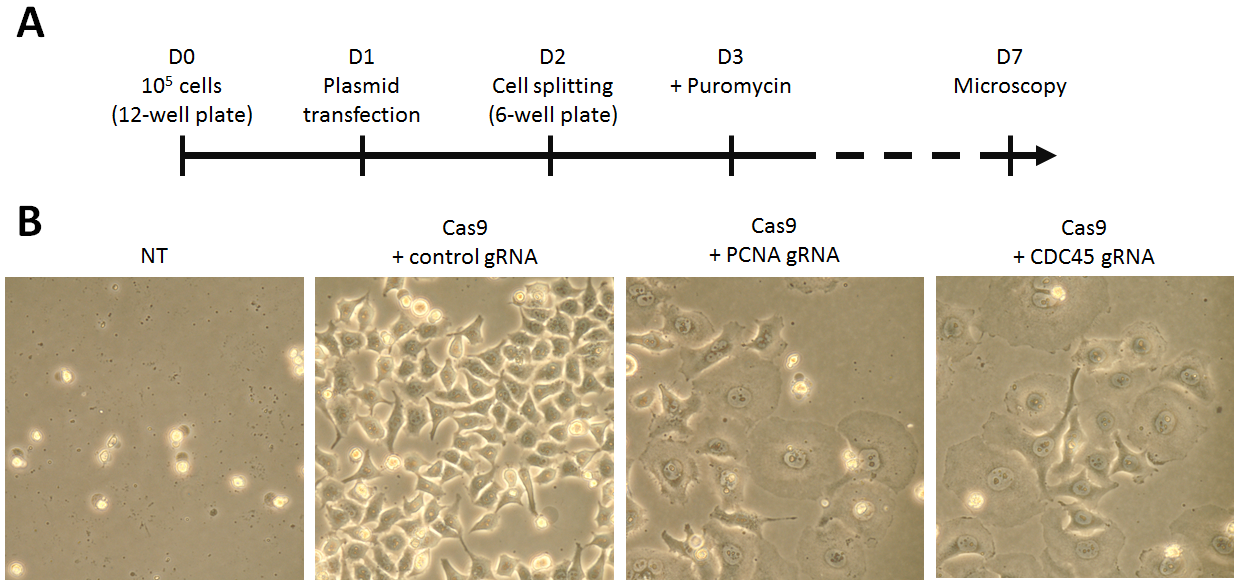
Fig 2. CRISPR/Cas9 gene disruption assay. (A) Schematic overview of the CRISPR/Cas9 assay. (B) Cell morphology after PCNA or CDC45 gene disruption with CRISPR/Cas9 technology. HeLa cells were transfected with pCMV-Cas9-gRNA plasmid using jetPRIME®. 48h post-transfection, the cells expressing Cas9 were selected using puromycin over 4 days. Cells transfected with empty vector are also shown (NT). The pictures show typical fields of the cells observed 6 days post-transfection at the same magnification (x400).
Co-expression of Cas9 and gRNA targeting either PCNA or CDC45 gene clearly affected the morphology and viability of puromycin-selected HeLa cells. Microscopy analysis six days post-transfection showed cell flattening and increased nuclear size in comparison to cells transfected with a plasmid expressing Cas9 and control gRNA. These phenotypical changes suggest efficient gene knock-out of PCNA and CDC45 gene achieved with the CRISPR/Cas9 genome editing method.
Conclusion & perspectives
Developing an easy and straightforward method for efficient delivery of one or multiple plasmid DNA coding for Cas9 and a gRNA of interest has become indispensable the in vitro use of CRISPR/Cas9 technology. Here, we demonstrated that the transfection reagent jetPRIME® is very efficient in delivering DNA plasmid encoding Cas9 and gRNA of PCNA or CDC45, using little amount of DNA while respecting cell morphology and viability. Moreover, jetPRIME® is suitable for multiple plasmid co-transfection which makes it the perfect solution for a variety of developed plasmid DNA-based CRISPR/Cas9 kits. Accordingly, several groups have already successfully used jetPRIME®-mediated transfection of the CRISPR/Cas9 system as illustrated by their respective publications (Langereis et al., 2015; Moore et al., 2015; Shi et al., 2015).
References
Desplancq, D., Freund, G., Conic, S., Sibler, AP., Didier, P., Stoessel, A., Oulad-Abdelghani, M., Vigneron, M., Wagner, J., Mély, Y., Chatton, B., Tora, L., Weiss, E. Exp Cell Res 342, 145-158 (2016).
Langereis, MA., Rabouw, HH., Holwerda, M., Visser, LJ., van Kuppeveld, FJ. J Virol 89, 11169-73 (2015).
Moore, R., Spinhirne, A., Lai, MJ., Preisser, S., Li, Y., Kang, T., Bleris, L. Nucleic Acids Res 43 1297-303 (2015).
Shi, J., Zhao, Y., Wang, K., Shi, X., Wang, Y., Huang, H., Zhuang, Y., Cai, T., Wang, F., Shao, F. Nature 526, 660-5 (2015).
Srinivasan, SV., Dominguez-Sola, D., Wang, LC., Hyrien, O., Gaultier. J. Cell Rep 3, 1629-39 (2013).